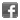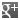Cookie Policy: This site uses cookies to improve your experience. You can find out more about our use of cookies in our Privacy Policy. By continuing to browse this site you agree to our use of cookies.
AdipoGen Life Sciences
SARS-CoV-2 Spike Protein S1 (RBD):Fc (human) (rec.) (B.1.617.2.1 Variant, Delta Plus)

| Product Details | |
|---|---|
| Synonyms | 2019-nCoV Spike Protein S1 (RBD); Spike Receptor Binding Domain; Delta Plus Variant; B.1.617.2.1; India; AY.1 |
| Product Type | Protein |
| Properties | |
| Source/Host | HEK 293 cells |
| Sequence |
Receptor-binding domain (RBD) of SARS-CoV-2 Spike protein S1 (aa 319-541) containing the mutations K417N, L452R & T478K is fused to the N-terminus of the Fc region of human IgG1. |
| Crossreactivity | Human |
| Biological Activity |
Binds to anti-SARS-CoV-2 Spike (RBD) antibodies in serum or plasma. Binds to human ACE2. |
| MW | ~60kDa (SDS-PAGE) |
| Purity | ≥95% (SDS-PAGE) |
| Endotoxin Content | <0.01EU/μg purified protein (LAL test). |
| Concentration | 1mg/ml after reconstitution. |
| Reconstitution | Reconstitute with 50µl endotoxin-free water. |
| Accession Number | QHD43416.1 |
| Formulation | Lyophilized. Contains PBS. |
| Other Product Data |
UniProt link QHD43416.1: SARS-CoV-2 S Protein |
| Shipping and Handling | |
| Shipping | BLUE ICE |
| Short Term Storage | +4°C |
| Long Term Storage | -20°C |
| Handling Advice |
After opening, prepare aliquots and store at -20°C. Avoid freeze/thaw cycles. Centrifuge lyophilized vial before opening and reconstitution. For maximum product recovery after thawing, centrifuge the vial before opening the cap. |
| Use/Stability |
Stable for at least 6 months after receipt when stored at -20°C. Working aliquots are stable for up to 3 months when stored at -20°C. |
| Documents | |
| MSDS |
 Download PDF Download PDF |
| Product Specification Sheet | |
| Datasheet |
 Download PDF Download PDF |
SARS-CoV-2 shares 79.5% sequence identity with SARS-CoV and is 96.2% identical at the genome level to the bat coronavirus BatCoV RaTG133, suggesting it had originated in bats. The coronaviral genome encodes four major structural proteins: the Spike (S) protein, Nucleocapsid (N) protein, Membrane/Matrix (M) protein and the Envelope (E) protein. The SARS Envelope (E) protein contains a short palindromic transmembrane helical hairpin that seems to deform lipid bilayers, which may explain its role in viral budding and virion envelope morphogenesis. The SARS Membrane/Matrix (M) protein is one of the major structural viral proteins. It is an integral membrane protein involved in the budding of the viral particles and interacts with SARS Spike (S) protein and the Nucleocapsid (N) protein. The N protein contains two domains, both of them bind the virus RNA genome via different mechanisms.
The CoV Spike (S) protein assembles as trimer and plays the most important role in viral attachment, fusion and entry. It is composed of a short intracellular tail, a transmembrane anchor and a large ectodomain that consists of a receptor binding S1 subunit (RBD domain) and a membrane-fusing S2 subunit. The S1 subunit contains a receptor binding domain (RBD), which binds to the cell surface receptor angiotensin-converting enzyme 2 (ACE2) present at the surface of epithelial cells. Recently, a new variant of SARS-CoV-2, called B.1.617 was detected in India. Three sublineages have been found, B.1.617.1 (variant Kappa) and B.1.617.3 containing 4 mutations in the Spike protein with a double mutations in the Receptor Binding Region (L452R, E484Q) and B.1.617.2 (variant Delta) that is different since it contains the mutation T478K instead of E484Q. These variants (especially the B.1.617.1 & B.1.617.2) of the SARS-CoV-2 coronavirus have evolved as fast-growing variants outspacing other variants. Recently, a new variant derived from Delta (called Delta Plus and containing the mutations K417N, L452R & T478K) was detected in India and in several other countries. While studies are still underway, scientists say Delta Plus does not seem to be more transmissible than Delta.