Cookie Policy: This site uses cookies to improve your experience. You can find out more about our use of cookies in our Privacy Policy. By continuing to browse this site you agree to our use of cookies.
Poly(ADP-ribosyl)ation (PARylation) – An Important Posttranslational Modification
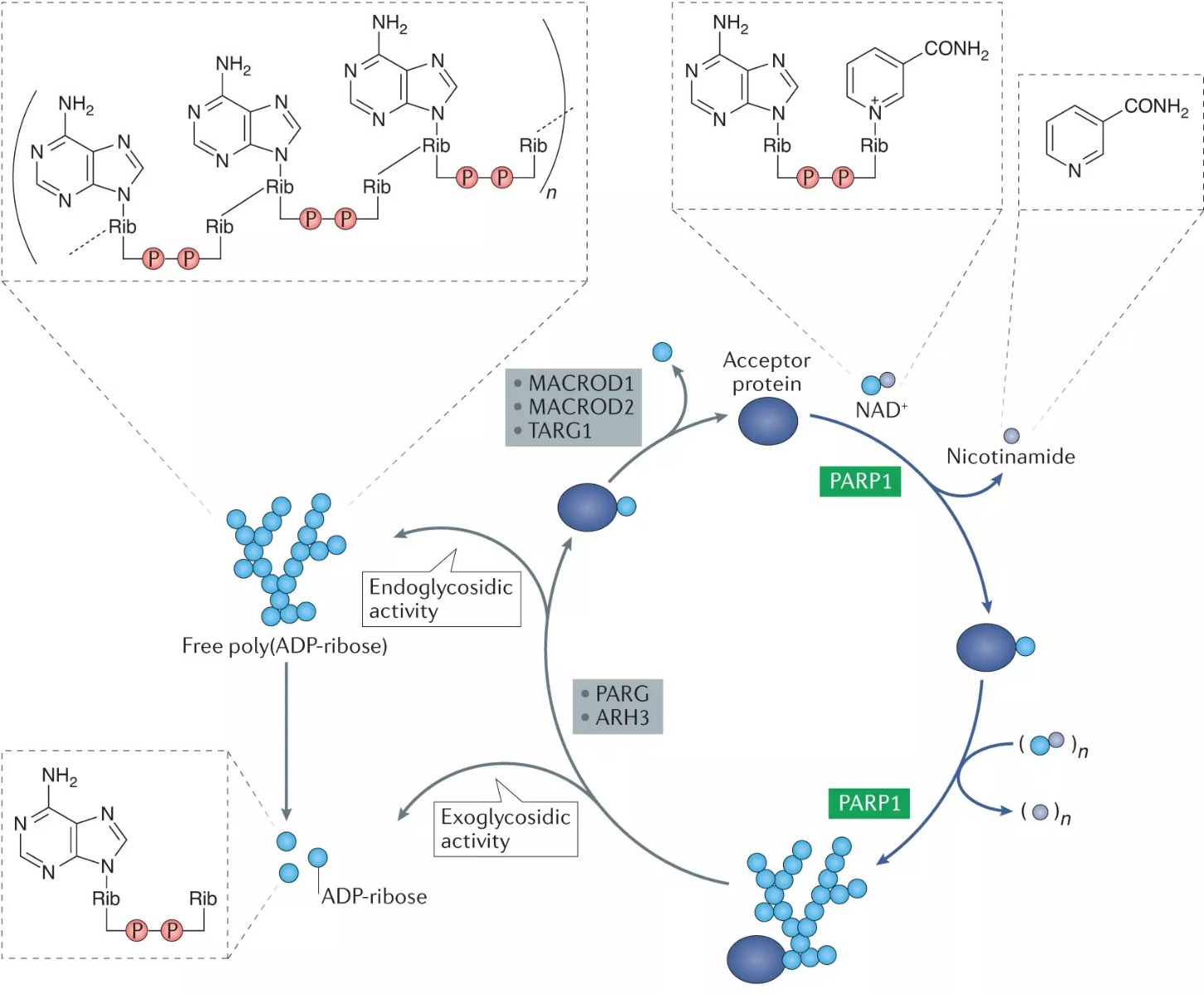
Poly(ADP-ribosyl)ation, also known as PARylation, is the post-translational modification process (similar to modifications like phosphorylation, methylation or acetylation) by which polymers of (ADP)-ribose (pADPr or PAR ranging from 2 to more than 200 ADP-ribosyl residues) are covalently attached to nuclear proteins by PAR polymerase (PARP) enzymes. The post-translational modification poly(ADP-ribosyl)ation (PARylation) plays key roles in cellular physiology and stress response. The modification is catalyzed by members of the family of poly(ADP-ribose) polymerases (PARPs, also known as ARTDs). It is involved in cellular functions including genome maintenance, DNA damage response, DNA repair, replication, transcription, chromatin remodeling, stress response, metabolism and regulation of cell death. In addition, PARPs represent promising targets in cancer therapy, either as chemosensitizers in combination with classical DNA damaging chemotherapeutics or as stand-alone drugs for tumors deficient in homologous recombination.
Figure: Schematic of PARylation and PAR removal (from N.J. Curtin & C. Szabo; Nat. Rev. Drug Discov. 19, 711 (2020)).
Poly(ADP-ribose) Polymerases (PARPs)
The posttranslational modification of nuclear proteins with poly(ADP)-ribose (PAR) is catalyzed for the most part by enzyme poly(ADP-ribose) polymerase 1 (PARP-1/ARTD1), and to a lesser extent by several additional poly(ADP-ribose) polymerases, by using NAD+ as substrate. PARP-1 is an abundant, highly conserved and constitutively expressed protein. Its catalytic activity, however, depends mainly on the presence of DNA single or double strand breaks, which are recognized by the two zinc fingers of the DNA-binding domain of the enzyme.
PARP-1 is selectively activated by DNA strand breaks to catalyze the addition of long branched chains of PAR to a variety of nuclear proteins, most notably PARP itself. The amount of PAR formed in living cells with DNA damage is commensurate with the extent of the damage. Under DNA damage conditions, PAR undergoes a rapid turnover, with a half-life in the range of minutes, as PAR is rapidly hydrolyzed and converted to free ADP-ribose by the enzyme poly(ADP-ribose)glycohydrolase (PARG).
Apart from automodified PARP-1, a number of proteins have been identified as target proteins (“acceptors”) for poly(ADP-ribosyl)ation, including the chromatin-architectural protein DEK. Importantly, PAR has been shown to bind with a variety of proteins in a non-covalent yet specific manner. Affinity of such non-covalent PAR interactions with specific binding proteins (i.e. XPA, p53, WRN) can be very high (nM range) and depends both on the PAR chain length and on the binding protein, which suggests the existence of a “PAR code”.
As indicated above, PAR regulates not only cell survival and cell-death programs, but also an increasing number of other biological functions with which novel members of the PARP family have been associated. These include transcriptional regulation, cell division, intracellular trafficking, inflammation and energy metabolism.
Literature References:
-
Functions of PARylation in DNA Damage Repair Pathways: H. Wei & X. Yu; Gen. Prot. Bioinform. 14, 131 (2016)
-
In Vivo Level of Poly(ADP-ribose): M. Miwa, et al.; Challenges 9, 23 (2018)
-
The Enigmatic Function of PARP1: From PARylation Activity to PAR Readers: T. Kamaletdinova, et al.; Cells 8, 1625 (2019)
-
Novel insights into PARPs in gene expression: regulation of RNA metabolism: Y. Ke, et al.; Cell. Mol. Life Sci. 76, 3283 (2019)
-
Poly(ADP-Ribose) Glycohydrolase (PARG) vs. Poly(ADP-Ribose) Polymerase (PARP) – Function in Genome Maintenance and Relevance of Inhibitors for Anti-cancer Therapy: D. Harrision, et al.; Front. Mol. Biosci. 7, 191 (2020)
-
Poly(ADP- ribose) polymerase inhibition: past, present and future: N.J. Curtin & C. Szabo; Nat. Rev. Drug Discov. 19, 711 (2020)
High Quality Reagents for PAR & PARP Research
The STANDARD anti-Poly(ADP-ribose) [PAR], mAb (10H) BULK Catalog Sizes Available!
A large part of the recent progress in the PARP field, especially as it relates to the catalytic activity of the enzymes, can be attributed to the development of novel ADP-ribose-detecting reagents. For decades, the field relies on the anti-PAR monoclonal antibody clone 10H (Prod. No. AG-20T-0001), which binds to PAR chains >10 ADP-ribose units in length and which even allows to visualize Poly(ADP-ribose) (PAR) formation at the level of intact cells by immunofluorescence. This anti-PAR (10H) antibody is therefore considered a standard antibody in the field of PARylation and has been published in hundreds of publications. This antibody has been used in Western blot, immunohistochemistry, immunocytochemistry and flow cytometry studies. The monoclonal antibody 10H recognizes poly(ADP-ribose) synthesized by a broad range of PARPs (poly(ADP-ribose) polymerases), including human, mouse, rat or Drosophila PARP enzymes.
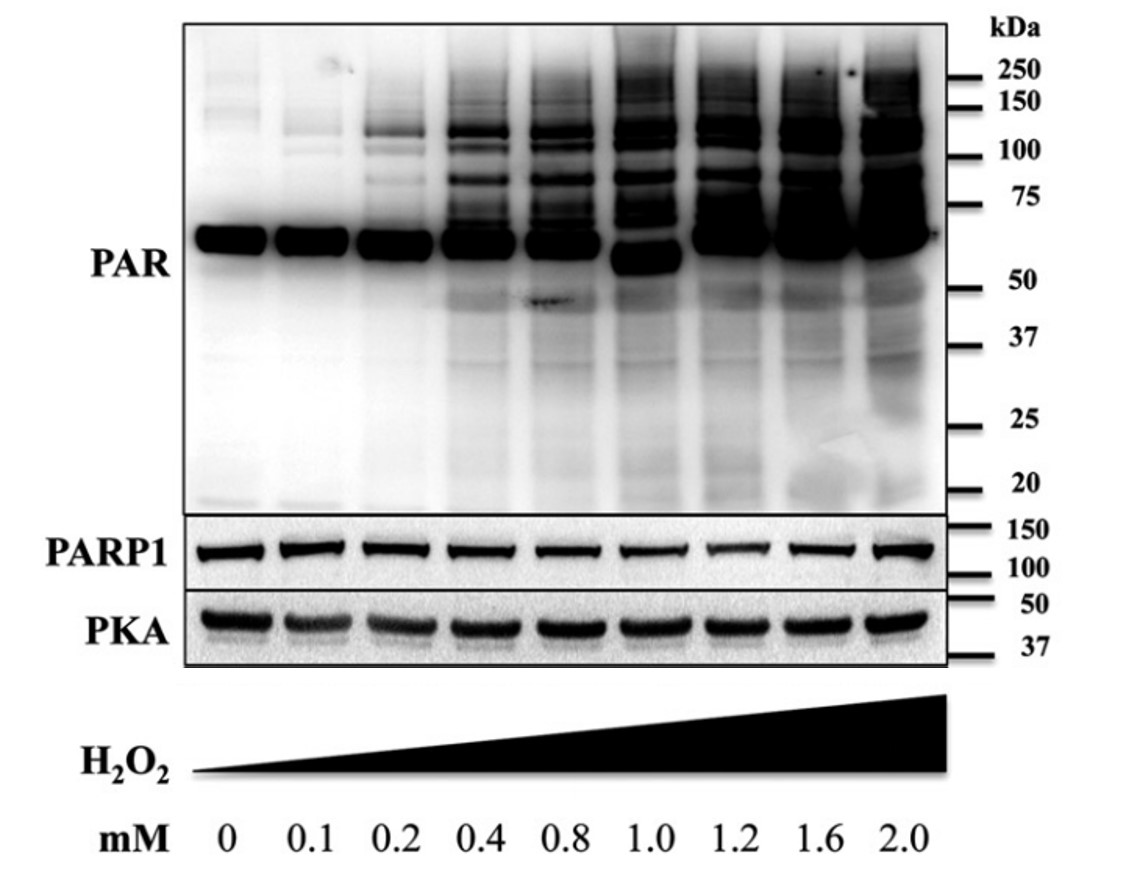
| Figure: Typical western blot pattern using anti-PAR (10H). Dose-dependence of H2O2 induced PARylation (PARP-1 activation) in U937 cells, as detected at 10min with a threshold of PARP-1 activation at 200mM. Control lanes show unchanged protein levels of PARP-1 and PKA under the same experimental conditions (A. Brunyanszki, et al.; Mol. Pharmacol. 86, 450 (2014)). |
 |
| Product Name | PID | Product Description |
| PARP-1 [ARTD1] (human) (rec.) (His) (high purity) | AG-40T-0011 | Human full-length PARP-1 [ARTD1] is fused to a MYC and His-tag. Produced in Sf21 cells. Activity: ≥1200U/mg protein. |
| PARP-2 [ARTD2] (mouse) (rec.) (His) (high purity) | AG-40T-0012 | Mouse full-length PARP-2 [ARTD2] is fused to a His-tag. Produced in Sf21 cells. Activity: ≥600U/mg protein. |
| PARP-3 [ARTD3] (human) (rec.) (His) (high purity) | AG-40T-0013 | Human full-length PARP-3 [ARTD3] is fused to a HA-tag and a His-tag. Produced in Sf21 cells. Activity: 10U/mg protein (only monoADP-ribosyl transferase activity). |
| PARP-1 [ARTD1] (E998K Mutant) (human) (rec.) Control | AG-40T-0015 | Human full-length inactive mutant E988K of PARP-1 is fused to a HA-tag and a His-tag. Produced in Sf21 cells. Activity: 0.5% of wild-type PARP-1. |
| UNIQUE PARG (human) (rec.) (His) (highly active) | AG-40T-0022 | Human PARG is fused to a His-tag. Produced in Sf21 cells. Activity: ~100ng is required for poly(ADP-ribose) degradation assays. |
| Product Name | PID | Product Description |
| anti-PARP-1 [ARTD1] (mouse), pAb | AG-25T-0001 | Recognizes mouse PARP-1 [ARTD1]. Cross-reacts weakly with human PARP-1. Works in ICC, IHC, IP and Western blot. |
| anti-PARP-1 [ARTD1] (human), pAb | AG-25T-0002 | Recognizes human PARP-1 [ARTD1]. Cross-reacts weakly with mouse and monkey PARP-1. Works in ICC, IHC, IP and Western blot. |
| anti-PARP-2 [ARTD2] (mouse), pAb | AG-25T-0003 | Recognizes mouse PARP-2 [ARTD2]. Cross-reacts weakly with human PARP-2. Works in ICC, IHC, IP and Western blot. |
| anti-PARP-10 [ARTD10] (human), mAb (5H11) | AG-20T-0004 | Recognizes human PARP-10 [ARTD10] with an epitope between aa 256-407, containing the glycine rich (G-rich) region. Works in ICC, IP and Western blot. |
| Product Name | PID | Product Description |
| AS-703026 | SYN-1190 | Selective, non-competitive, orally bioavailable MEK1/2 inhibitor, shown to induce apoptosis via caspase-3 and PARP cleavage in multiple myeloma cells. |
| AZ9482 | SYN-3046 | Potent and selective PARP inhibitor (IC50s = 1, 1, 46, 640, 9, and 160 nM for PARP-1, -2, -3, -6, TNKS1/PARP-5a, and TNKS2, respectively). |
| Benadrostin | BVT-0079 | Rare natural product PARP inhibitor (IC50=35μM). |
| DPQ | AG-CR1-0037 | Cell permeable potent PARP-1 inhibitor (IC50=40 nM). It is ~10-fold less potent against PARP-2. |
| GeA-69 | AG-CR1-3706 | Potent, selective, allosteric inhibitor of macrodomain 2 of poly-adenosine-diphosphate-ribose polymerase 14 (PARP-14 M2) (Kd=0.86µM). PARP-14 is an anti-apoptotic pro-survival protein associated with inflammatory diseases and several types of cancer. |
| Genipin | AG-CN2-0481 | Induces PARP cleavage and caspase-3- and caspase-9-mediated apoptosis. |
| 1,5-Isoquinolinediol | AG-CR1-0024 | Cell permeable potent PARP-1 inhibitor (IC50=0.39μM). |
| IWR-1-endo | AG-CR1-3581 | Cell permeable Tankyrase-1/PARP-5a (IC50=131nM) and Tankyrase-2/PARP-5b (IC50=56nM) inhibitor (in vitro auto-PARylation assays). |
| MK-4827 tosylate | CDX-M0565 | Orally bioavailable inhibitor of PARP-1 and PARP-2 (IC50s=3.8 and 2.1nM, respectively). |
| Olaparib | CDX-O0144 | Cell permable potent inhibitor of PARP-1 and PARP-2 (IC50=5 and 1nM, respectively). Less effective against the Tankyrase-1/PARP-5a (IC50=1.5μM). |
| PJ-34 | AG-CR1-0100 | Cell permeable and water soluble potent PARPs inhibitor (EC50=20nM). It also binds and inhibits the Tankyrase-1/PARP-5a (IC50=1μM). |
| PARG Inhibitor PDD00017273 | AG-CR1-3646 | Potent PARG inhibitor. Inhibits human recombinant PARG enzymes (IC50=26nM) and in cell assays (IC50=37nM). Inactive against ARH3 and PARP-1s (IC50>30µM). |
