Cookie Policy: This site uses cookies to improve your experience. You can find out more about our use of cookies in our Privacy Policy. By continuing to browse this site you agree to our use of cookies.
AdipoGen Life Sciences
MTSSL
As low as
90
CHF
CHF 90.00
In stock
Only %1 left
AG-CR1-3573-M01010 mgCHF 90.00
AG-CR1-3573-M05050 mgCHF 360.00
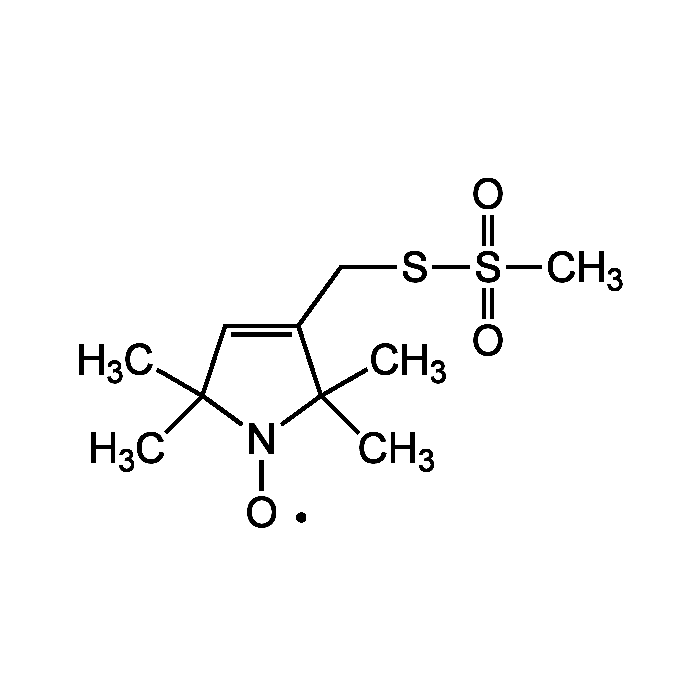
| Product Details | |
|---|---|
| Synonyms | Mts-SL; MTSL; (1-Oxyl-2,2,5,5-tetramethylpyrroline-3- methyl)methanethiosulfonate; Otmpmms |
| Product Type | Chemical |
| Properties | |
| Formula |
C10H18NO3S2 |
| MW | 264.3 |
| CAS | 81213-52-7 |
| Purity Chemicals | ≥98% (HPLC) |
| Appearance | Yellow crystalline solid. |
| Solubility | Soluble in ethanol, methanol, DMSO, acetonitrile or acetone. *See below for solubility in aqueous solutions. |
| Other Product Data |
*Solubility in aqueous solutions: MTSSL can be used in aqueous solution at concentrations as high as 10mM. However, MTSSL initially must be dissolved in an organic solvent such as DMSO or acetonitrile (100mM or higher) and then the initial stock solution can be diluted 10-100 fold in water-based buffers. Sonication and heating is not recommended to dissolve MTSSL. |
| InChi Key | BLSCGBLQCTWVPO-UHFFFAOYSA-N |
| Smiles | CC1(C)C=C(CSS(C)(=O)=O)C(C)(C)N1[O] |
| Shipping and Handling | |
| Shipping | BLUE ICE |
| Short Term Storage | +4°C |
| Long Term Storage | -20°C |
| Handling Advice | Protect from light. |
| Use/Stability | Stable for at least 2 years after receipt when stored at -20°C. |
| Documents | |
| MSDS |
 Download PDF Download PDF |
| Product Specification Sheet | |
| Datasheet |
 Download PDF Download PDF |
Description
- Highly reactive, thiol-specific spin-label.
- Specific conformational probe of thiol site structure by its minimal rotational freedom and distance from the covalent disulfide linkage to the macromolecule under study.
- Used to label cysteine residues in proteins (site-directed labeling, SDS-labeling).
- Allows protein structure and protein dynamics determination as well as the study of protein-protein and protein-oligonucleotide interactions.
Product References
- A novel reversible thiol-specific spin label: papain active site labeling and inhibition: L.J. Berliner, et al.; Anal. Biochem. 119, 450 (1982)
- ESR spectra reflect local and global mobility in a short spin-labeled peptide throughout the alpha-helix-coil transition: A.P. Todd & G.L. Millhauser; Biochemistry 30, 5515 (1991)
- Kinetics and motional dynamics of spin-labeled yeast iso-1-cytochrome c: 1. Stopped-flow electron paramagnetic resonance as a probe for protein folding/unfolding of the C-terminal helix spin-labeled at cysteine 102: K. Qu, et al.; Biochemistry 36, 2884 (1997)
- Pressure-induced thermostabilization of glutamate dehydrogenase from the hyperthermophile Pyrococcus furiosus: M.M. Sun, et al.; Protein Sci. 8, 1056 (1999)
- Protein global fold determination using site-directed spin and isotope labeling: V. Gaponenko, et al.; Protein Sci. 9, 302 (2000)
- High-resolution probing of local conformational changes in proteins by the use of multiple labeling: unfolding and self-assembly of human carbonic anhydrase II monitored by spin, fluorescent, and chemical reactivity probes: P. Hammarstroem, et al.; Biophys. J. 80, 2867 (2001)
- Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme: P.P. Borbat, et al.; JACS 124, 5304 (2002)
- Methods for study of protein dynamics and protein-protein interaction in protein-ubiquitination by electron paramagnetic resonance spectroscopy: H.J. Steinhoff; Front. Biosci. 7, c97 (2002)
- Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein-protein and protein-oligonucleotide interaction: H.J. Steinhoff; Biol. Chem. 385, 913 (2004)
- Dynamics of the nitroxide side chain in spin-labeled proteins: F. Tombolato, et al.; J. Phys. Chem. B. 110, 26248 (2006)
- Parametrization, molecular dynamics simulation, and calculation of electron spin resonance spectra of a nitroxide spin label on a polyalanine alpha-helix: D. Sezer, et al.; J. Phys. Chem. B. 112, 5755 (2008)
- Potential artifacts in using a glutathione S-transferase fusion protein system and spin labeling electron paramagnetic resonance methods to study protein-protein interactions: C. Antoniou & L.W. Fung; Anal. Biochem. 376, 160 (2008)
- Alzheimer’s Aβ42 and Aβ40 form mixed oligomers with direct molecular interactions: L.Gu & Z. Guo; BBRC 534, 292 (2021)
- Lipid membranes induce structural conversion from amyloid oligomers to fibrils: L. Gu & Z. Guo; BBRC 557, 122 (2021)
- Static and dynamic disorder in Aβ40 fibrils: H. Xiao, et al.; BBRC 610, 107 (2022)




![N-tert-Butyl-alpha-phenylnitrone [PBN]](https://adipogen.com/media/catalog/product/cache/60eb5af712bc93baae8d55513bd31b01/i/m/image_670.png)